Please… Draw me a project
Maëlle Salmon 🏠 https://masalmon.eu 🐦 ma_salmon
Licence CC-BY-SA
👋 Hello from Nancy!

Picture by Dimitry Anikin on Pexels.
I’m delighted to attend satRday Nairobi. It is my fourth satRday conference!
satRday Cape Town, 2018
satRday Cardiff, 2018

satRday Paris, 2019

Today:
Managing your R project
I will share about some tools and tips!

Picture by cottonbro on Pexels.
But first, who am I?
Recent work
Part-time Research Software Engineer for rOpenSci, in particular maintaining our package dev guide. 🔧
Work on the next version of pkgdown. ✨
Online book “HTTP testing in R” with Scott Chamberlain. 📖
Course “Scientific Rmd blogging”. 📝
Current volunteering
R-Ladies Global Twitter Account. 🐦
Editor for rOpenSci Software Peer Review. 📦
Various talks. 😉
My slidedeck is online 😁
Goals of my talk
That everyone learns at least one new thing. 😁
⚒️ Basic principles for R projects ;
⚒️ How to protect your projects from external changes;
⚒️ What structure for your project ;
⚒️ How to run your project.
Why give advice on projects?
😿 Not asked for by The Little Prince.
😺 Improve the life of anyone touching or reading your results and their origin. Reproducibility.
😹 Always something to improve or procrastinate on.
Why listen to me
on this topic?
😦 Not regularly in charge of analyses.
😄 I keep up-to-date with R news.
Some basics

Picture by cottonbro on Pexels.
Main source of advice : Jenny Bryan!
“Everything I know is from @JennyBryan.” —@sharlagelfand #mood #rstudioconf
— Kara Woo (@kara_woo) January 30, 2020
As an aside, Sharla Gelfand’s presentation
Don’t repeat yourself, talk to yourself! Repeated reporting in the R universe
Sharla too is a great source of good ideas! 💐
Main source of advice : Jenny Bryan!

Logo by Martin Monkman.
Your project, a garden?

Picture by Irina Iriser on Pexels.
Yes, but an independent unit!
A project, a folder!
The Kei-tora Gardening Contest is an annual event put on by the Japan Federation of Landscape Contractors to see who can build the best looking garden on the back of a Kei truck. Here are some of the winners from the past 12 years. https://t.co/IpkRiALn7a pic.twitter.com/vep4K0VZt6
— Doctor Popular 💉 💉 🎉 (@DocPop) June 10, 2021
🔥
If the first line of your script is …
🔥 setwd("C:/users/you/a/path/specific/to/you);
🔥 or rm(list = ls());
Jenny Bryan will come into your office and set your computer on fire. 😱
File paths relative
to the project root
NO:
readr::read_csv("/home/maelle/Documents/conferences/satRday_nairobi/data/cool-stuff.csv")
File paths relative
to the project root

Illustration by Allison Horst.
Package here
by Kirill Müller
-
.Rprojor an empty file called.hereat the project root. -
Paths are defined relative to that.
here::here()
[1] "/home/maelle/Documents/conferences/satRday_nairobi"
here::here("data")
[1] "/home/maelle/Documents/conferences/satRday_nairobi/data"
Package here
by Kirill Müller
readr::read_csv(here::here("data", "cool-stuff.csv"))
It works from anywhere inside my satRday_nairobi folder! (satRday_nairobi/README.Rmd, satRday_nairobi/reports/script.R, etc.)
Always start fresh
-
Re-start R regularly and without fear!
-
NEVER save and re-load
.RData!
`usethis::use_blank_slate()` sets your @rstudio preference to NEVER save/restore .RData on exit/startup, which is a lifestyle endorsed by many #rstats folks (including me).
— Jenny Bryan (@JennyBryan) June 15, 2021
Just did a clean install and got my first chance to use this on my own behalf 😌https://t.co/Qwd8VzaCVn
“Project-oriented workflow”
🌹 A way of life, hum, work!
✨ usethis::create_project() ✨
The name of your roses
That which we call a rose by any other name would smell as sweet
Shakespeare in Romeo and Juliet. 🌹
Not true when writing code! 😅
Naming your files
-
Machine readable (no accent);
-
Human readable (informative about the content);
-
Work well with default ordering (YYYY-MM-DD rather than DD-MM-YYYY).
Naming things
I didn't expect programming to involve so much time studying a thesaurus #namingThings
— Jenny Bryan (@JennyBryan) September 1, 2021
Your project over time
🌻 BACKUP! 🌻

Picture by Skitterphoto on Pexels.
Version control
Your project will evolve, how to keep track of changes to be able to come back?

Picture by PhotoMIX Company on Pexels.
Version control
-
Dates in filenames
-
or version control.
Learning git: not easy, but worth the effort! Not R but useful for R.
Being able to try things out, come back, understand past changes.
What is git?
![[Person 1 points to a computer on a desk while two other people are standing further away behind an office chair.] Person at the computer: This is git. It tracks collaborative work on projects through a beautiful distributed graph theory tree model. Other person: Cool. How do we use it? Person at the computer: No idea. Just memorize these shell commands and type them to sync up. If you get errors, save your work elsewhere, delete the project, and download a fresh copy..](https://imgs.xkcd.com/comics/git.png)
XKCD by Randall Munroe.
What is git?
Drawing by Damien Cornu. ❤️
Some git commands roughly explained
-
git addto start tracking a file (.gitignoreto list files to always ignore) ; -
git committo save a change in the history ; -
git pull/git pushto download / upload the local version from / to the remote version (e.g. GitHub); -
git checkout -bto create a “branch.”
Where to use
git commands?
My preferences 😁
- Packages usethis (
usethis::use_git(),usethis::use_github(), etc.) and gert (gert::git_push()) to stay in R ; - git window in RStudio to click not too far away from R ;
- Command line for commands copy-pasted from Stack Overflow ;
- A graphical interface for better visualization? E.g. GitKraken, VSCODE.
Resources about git
-
Excuse Me, Do You Have a Moment to Talk About Version Control?, Jenny Bryan.
-
Happy Git and GitHub for the useR, Jenny Bryan, the STAT 545 TAs, Jim Hester.
-
Reflections on 4 months of GitHub: my advice to beginners, Suzan Baert.
Until now, what we learnt from Jenny Bryan…
-
Isolate R projects, re-start R regularly.
-
Name files well.
-
Use version control.
More wisdom
by Jenny Bryan et al
🌹 Good enough practices in scientific computing Wilson G, Bryan J, Cranston K, Kitzes J, Nederbragt L, et al. (2017) Good enough practices in scientific computing. PLOS Computational Biology 13(6): e1005510. https://doi.org/10.1371/journal.pcbi.1005510
🌹 What They Forgot to Teach You About R, Jenny Bryan, Jim Hester.
Protect your project from external changes

Picture by cottonbro on Pexels.
A scary story…
-
You write beautiful data wrangling with
package::my_favorite_function()… -
Now you go and update that package and realize
my_favorite_function()is gone!
For good reasons but your script is now broken! 😱
Store project dependencies
Encapsulate your project! 🎉
Important tool : renv by Kevin Ushey!
Successor of packrat.
renv in three steps
-
renv::init() -
Install and remove packages as usual. Regularly
renv::snapshot(). Metadata of dependencies stored inrenv.lock. 🔒 -
Your colleague who inherits your project runs
renv::restore().
Even more encapsulating
🐳 Docker? 🔒 R version, operating system, in short everything.
-
“Introduction to using Docker for reproducibility in R” by Malindrie Dharmaratne at R-Ladies Brisbane. Video recording, materials.
Protect your project from external changes
List dependencies of the project.
The easiest way to get started is to start using renv.
What file structure
for your project?

Picture by cottonbro on Pexels.
What’s in your project?
-
Data or the code to get them from a database or a remote resource;
-
Some code munging and analysing them;
-
Some output that could be a graph, a report etc.
What file structure
for your project?
-
Consistent.
-
Automatic creation.
-
… Package or not?
projectTemplate
By Kenton White.
Love for ProjectTemplate, Hilary Parker:
✅ “Routine is your friend.”
✅ “It’s easier to start somewhere and then customize, rather than start from the ground up.”
✅ “Reproducibility should be as easy as possible.”
✅ “Finding things should also be as easy as possible.”
Make an R package?
-
To store functions and data used across projects: YES!!!
E.g. follow Shel Kariuki’s tutorial given at R-Ladies Nairobi. 😉
- To e.g. store an RStudio project template or re-create a folder structure: YES!!!
- To store your project i.e. a project as a package : maybe.
Analysis project = 📦
-
Dependencies in
DESCRIPTION, -
Functions in
R/with documentation inman/, -
Data in
data/(ordata-raw/), -
Analyses as vignettes (R Markdown),
-
Informative README.
Analysis project = 📦?
🏄♂️ Re-use or refresh your package development skills,
🏄♂️ Re-use tools made for package development (like devtools and usethis).
“Research compendium.” Packaging Data Analytical Work Reproducibly Using R (and Friends), Ben Marwick, Carl Boettiger & Lincoln Mullen (2018), The American Statistician, 72:1, 80-88, DOI: <10.1080/00031305.2017.1375986>
Project as package, specific tools
📦 rrtools by Ben Marwick. Set up a compendium!
📦 holepunch by Karthik Ram. One click and the reader gets to play with your code! (🤫 holepunch works without the compendium structure as well.)
🚀 R-universe by Jeroen Ooms at rOpenSci to publish your analyses.
Project as a package: no?
Project as an R package: An okay idea by Miles McBain.
“My response to advocates of project as a package is: ==You’re wasting precious time making the wrong packages.==”
“Instead of shoehorning your work into the package development domain, with all the loss of fidelity that entails, why aren’t you packaging tools that create the smooth {devtools}/{usethis} style experience for your own domain?”
What file structure
for your project?
As you wish 😉 (as your team wishes) but
-
Basic structure consistent over time ;
-
Automatic creation.
Make reproducibility ✨ easier ✨.
How to run
your project?

Picture by cottonbro on Pexels.
Running the project?
How do you go from resources and scripts to the analysis output (e.g. report, figures)?
🤸
How to run
your project?
Maybe you only need the knit button if your project is an R Markdown report?
Maybe you need something more complex?
Let’s discuss two cases
-
Optimize a pipeline.
-
Track versions of an analysis (input and output).
Let’s discuss two cases
-
Optimize a pipeline. 📦 targets maintained by Will Landau.
-
Track versions of an analysis (input and output). 📦 orderly maintained by Rich FitzJohn.
targets by Will Landau
-
targets deduces relationships between pieces of a project (e.g. if raw data changes, everything needs to be re-done) ;
-
targets only performs necessary computation.
Part of the rOpenSci suite of packages. Successor of drake by the same author.
targets
At the core of a targets project, the _targets.R file.
-
Load packages;
-
Load functions (
source()scripts fromR/for instance); -
Define targets!
Targets
list(
tar_target(
raw_data_file,
"data/raw_data.csv",
format = "file"
),
tar_target(
raw_data,
read_csv(raw_data_file, col_types = cols())
),
tar_target(
data,
raw_data %>%
filter(!is.na(Ozone))
),
tar_target(hist, create_plot(data)),
tar_target(fit, biglm(Ozone ~ Wind + Temp, data))
)
How does it run?
-
To build,
targets::tar_make()(andtargets::tar_destroy()); -
To understand your pipeline,
targets::tar_glimpse()&co.
Visualizing the pipeline
targets::tar_glimpse()
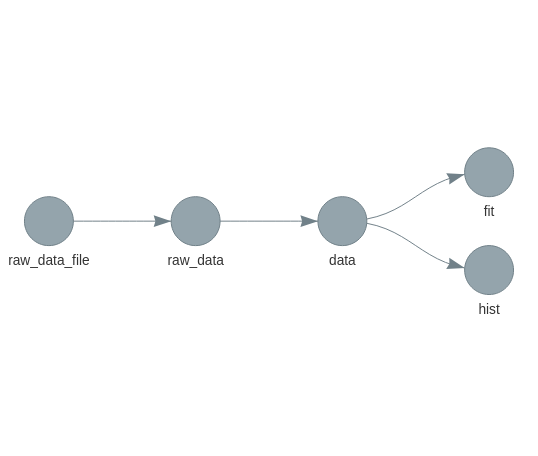
Illustration from targets manual.
How to get started with targets?
-
Reproducible Computation at Scale in R with {targets} (Will Landau at RUG Lille).
-
Start with a small project (My current level 😅).
The R Targetopia
An R package ecosystem for democratized reproducible pipelines at scale
How to keep up with targets?
-
Watch the GitHub repository of targets ;
-
Follow Will Landau on Twitter ;
-
Subscribe to rOpenSci newsletter ;
-
Connect with other users. 😉
orderly by Rich FitzJohn
(Thanks to Rich for answering my questions 🙏)
Different challenge: keep track of everything going into and out of an analysis, at different points in time.
For instance to allow audits in the future.
The analysis can be huge so git isn’t the tool for the job.
orderly
In orderly there are repos and in repos there are reports.
Example with a repo of one report. orderly::orderly_init("blop") then orderly::orderly_new("example", "blop"), some file editing and I get:
blop
├── orderly_config.yml
└── src
└── example
├── orderly.yml
└── script.R
Configuration src/example/orderly.yml
script: script.R
artefacts:
- staticgraph:
description: A graph of things
filenames: mygraph.png
- data:
description: Data that went into the plot
filenames: mydata.csv
Example script
dat <- data.frame(x = 1:10, y = runif(10))
write.csv(dat, "mydata.csv", row.names = FALSE)
png("mygraph.png")
plot(dat)
dev.off()
How does it run?
- Experiment with the “development mode”.
- Create a draft version.
id <- orderly::orderly_run("example", root = "blop")
This draft (resources, script, results, in short everything!) appears in the folder draft/example/id-illisible.
- If happy, “commit.”
orderly::orderly_commit(id, root = "blop")
This version (resources, script, results, in short everything!) appears in the folder archive/example/id-illisible.
How does it run?
⚠️The archive and draft folders can be huge, so back them up with something other than git. ⚠️
How to get started with orderly?
-
orderly documentation website is really great!
-
Start small to understand how it works (once again, my level 👋).
-
Connect with other users.
How to keep up with orderly?
-
Watch orderly GitHub repository ;
-
Follow the blog of the team developing orderly ;
-
Follow Rich FitzJohn on Twitter.
How to run
your project…
Conclusion

Picture by cottonbro on Pexels.
Thank you
Before summing up, thanks to the organization team and to all of you! 🙏 ✨
Thanks a lot to Christophe Dervieux for useful feedback on the content of this talk!
How to draw a project?
🌻 Good basics like isolating your project, back-ups.
🌻 Encapsulating the project. (renv? Docker?)
🌻 Practical, consistent and automatic structure. (Package or not?)
🌻 Using tools for building outputs that answers your needs (optimizing a pipeline? tracking versions of an analysis projects?).
How to learn how to draw a project?
Some more useful resources:
-
Course “Reproducible Research Data and Project Management in R” by Anna Krystalli.
-
Good enough practices in scientific computing Wilson G, Bryan J, Cranston K, Kitzes J, Nederbragt L, et al. (2017) Good enough practices in scientific computing. PLOS Computational Biology 13(6): e1005510. https://doi.org/10.1371/journal.pcbi.1005510
-
The Turing Way, an open source community-driven guide to reproducible, ethical, inclusive and collaborative data science.
How to draw a project?
🌹 Read everything Jenny Bryan wrote.
🌹 Choose or even create, as a team, the box in which you put and build your project.
🌹 Do not be afraid to renew your toolset over time.